In the case of 2D NMR, this operation is more tedious as it has to be applied to both dimensions. In addition, the low digital resolution typically obtained in 2D NMR (nD in general) spectra makes calibration more sensitive to the actual position of the reference peak selected for referencing.
Most of the NMR software packages I know include facilities to manually align the 2D and 1D spectra, that is, to correlate signals of 2D and 1D spectra. However, I was interested in a fully automatic procedure to take advantage of the higher digital resolution of the 1D spectrum to calibrate the 2D spectrum.
After studying this issue in depth, we came out with a new algorithm which works exactly as I wanted: Once the 1D spectrum is properly referenced, the 2D spectrum is automatically calibrated with the information contained in the 1D spectrum.
As always, a picture is worth a thousand words. Consider the following 2D COSY spectrum:
And compare it with the corresponding high resolution 1H-NMR counterpart:
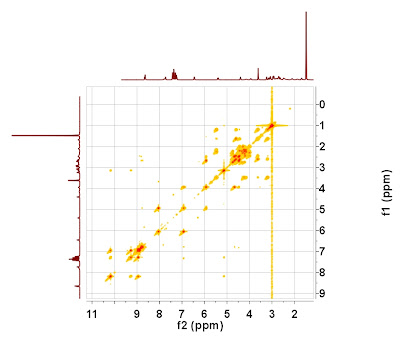
It can be observed that the chemical shift scales of both spectra differ by about 1.5 ppm. If the 1D spectrum is attached to the 2D COSY as an external projection, the result would look something like this:
Application of the newly developed autoalignment algorithm will yield the following result (please note that there is not need to enter any user-defined parameter: it's fully automatic):
The scales of the 2D spectrum have been calibrated by using the information contained in the high resolution 1H spectrum.
It’s important to mention that at present, this algorithm does not work with all 2D spectra. Basically, it works fine in those spectra in which internal projections are compatible, in terms of number of resonances, with the external 1D traces. This would be the case of 2D homonuclear experiments and the proton dimension of HSQC and related spectra, but it will not work in the